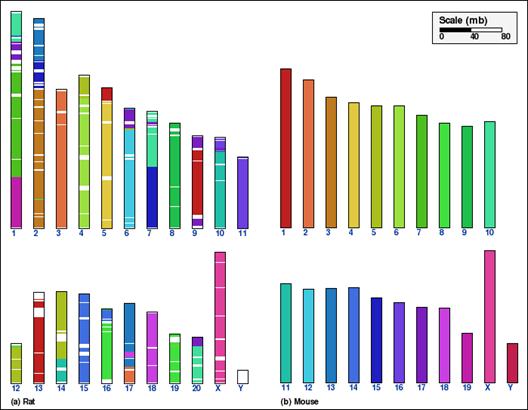
Whole genome comparison for two pairs of mammalian genomes, using alternative aggregation approaches. In panels A and B, the intermediate aggregation (INTAGG) is used, leading to reversal distances of 21 and 128, respectively. In panels C and D, the same setup of parameters is used, except that only orthologs common to five mammalian genomes, as specified in the text, are used. In the latter case, reversal distances of 14 and 86, respectively, are obtained.
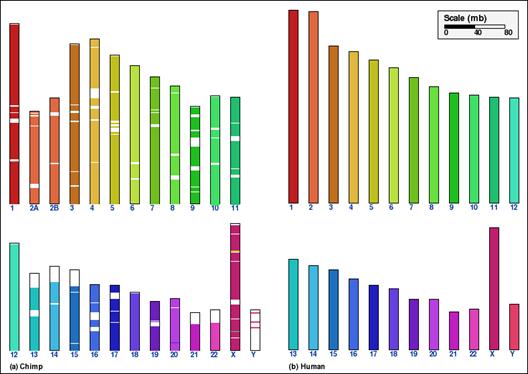
Panel B
Panel C
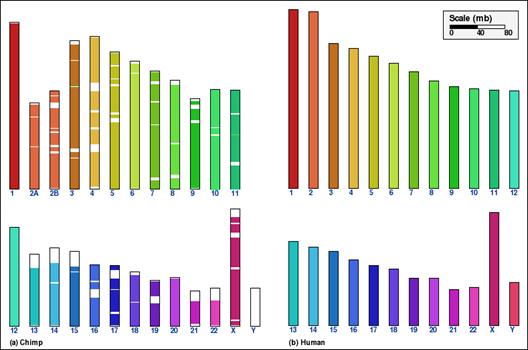
Panel D
Chromosome level effect of using multiple genomes
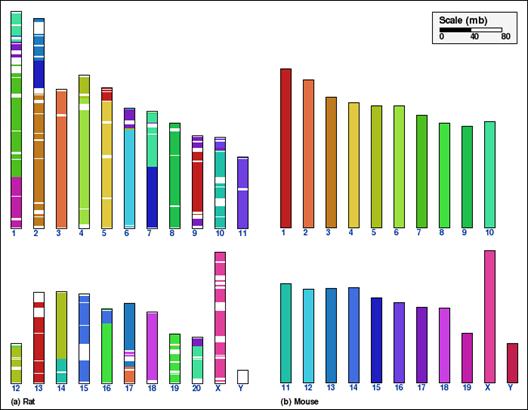
This example shows analysis of synteny for X chromosome of mouse and rat genomes. Panel A shows the synteny between the two chromosomes without aggregation and orthologs identified for each pair of genomes independently are used. Panel B shows the default coarse-graining (DEFAGG), with the same subset of orthologs. Finally, panel C shows the result of no aggregation, however, with a set of orthologs common to five mammalian genomes (human, mouse, rat, dog, chimpanzee).
Comparison of the number of synteny blocks, RD as well as visual comparison makes it evident that the using multiple genomes allows natural aggregation inspite of setting the parameters to zero. This provides a mechanism for overcoming the effect of the parameters.
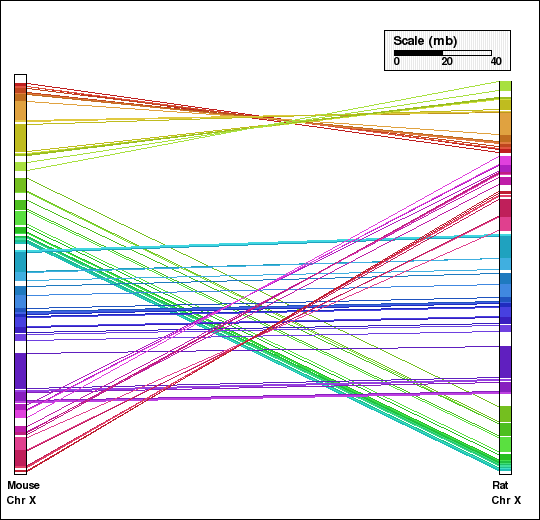
Panel E: Comparison of X chr of mouse and rat genomes using NOAGG. Number of synteny blocks is 49 and RD is 14.
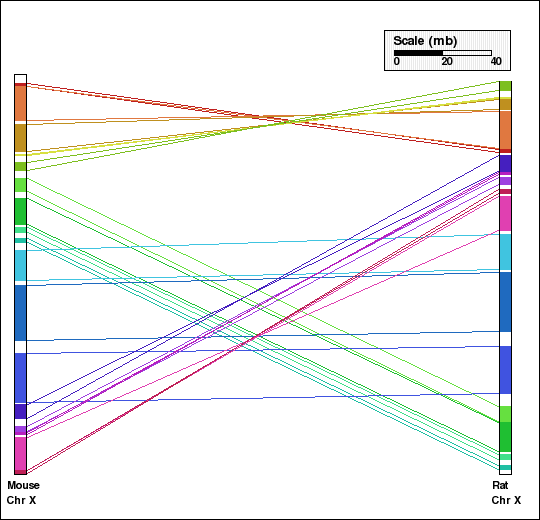
Panel F: Comparison of X chr of mouse and rat genomes using DEFAGG. Number of synteny blocks is 17 and RD is 8.
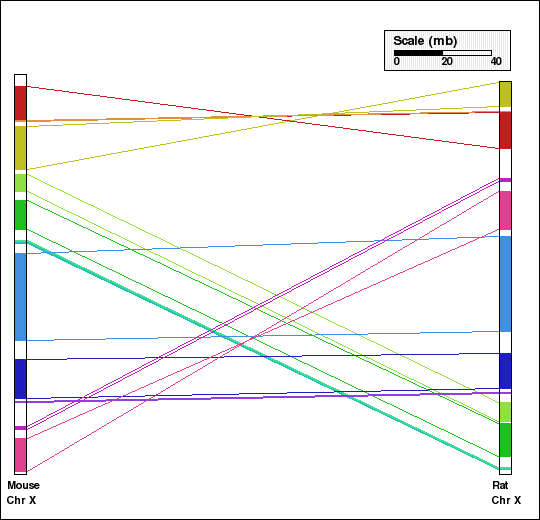
Panel G: Comparison of X chr of mouse and rat genomes using NOAGG with 5-way approach (human, mouse, rat, dog and chimpanzee genomes). Number of synteny blocks is 12 and RD is 6.